Introducing Dolt + Metaflow
Note: this blog post appeared on previously on Medium and represents a collaboration with the Metaflow maintainers at Netflix.
Background
This post details how to use Metaflow with Dolt. Metaflow is a framework for defining data science and data engineering workflows with the the ability to define local experiments and scale those experiments to production jobs from a single API. Dolt is a version controlled relational database. It provides a familiar SQL interface along with Git-like version control features. Each commit corresponds to the state of the database at the time the commit was created. Both Dolt and Metaflow are open source.
Specifically we show how the combination of Dolt and Metaflow can be used to address three common challenges in modern data science and data engineering settings:
- Reproducibility: how to make sure that we can repeat past results consistently and improve results incrementally. Disciplined science and engineering requires that goalposts don’t shift unexpectedly, for example due to changes in input data.
- Experiment tracking - how to keep track of all changes and experiments executed. Today, it is hard to imagine software development without a version control like Git - data science needs a similar tool to stay organized.
- Lineage and auditing - finally, when our system is in production, we want to understand the exact model and input data that contribute to the system’s output, for example predictions. This is especially important when results are surprising and we need to understand why.
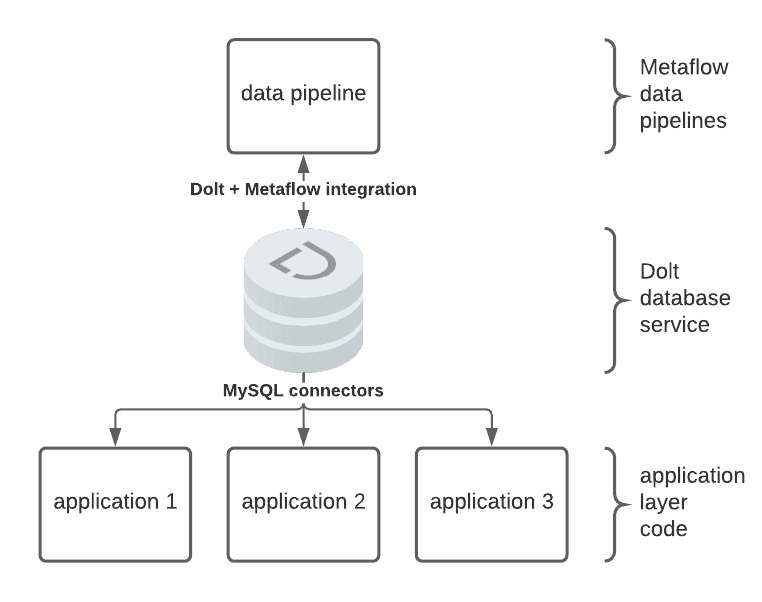
Both Dolt and Metaflow are built around the idea of strong versioning: Dolt versions data and Metaflow versions code, execution environments, and the state of execution. To illustrate how Metaflow and Dolt work together to solve for these challenges, we use an example Metaflow pipeline to derive results data from an input dataset. We read the input data into a Pandas DataFrame and convert that DataFrame to permanent storage in Dolt for use with application layer services with an intermediate step. Dolt SQL Server can be used to serve this versioned and reproducible data over MySQL connectors:
Let's dive into the details of our example pipeline.
Example Pipeline
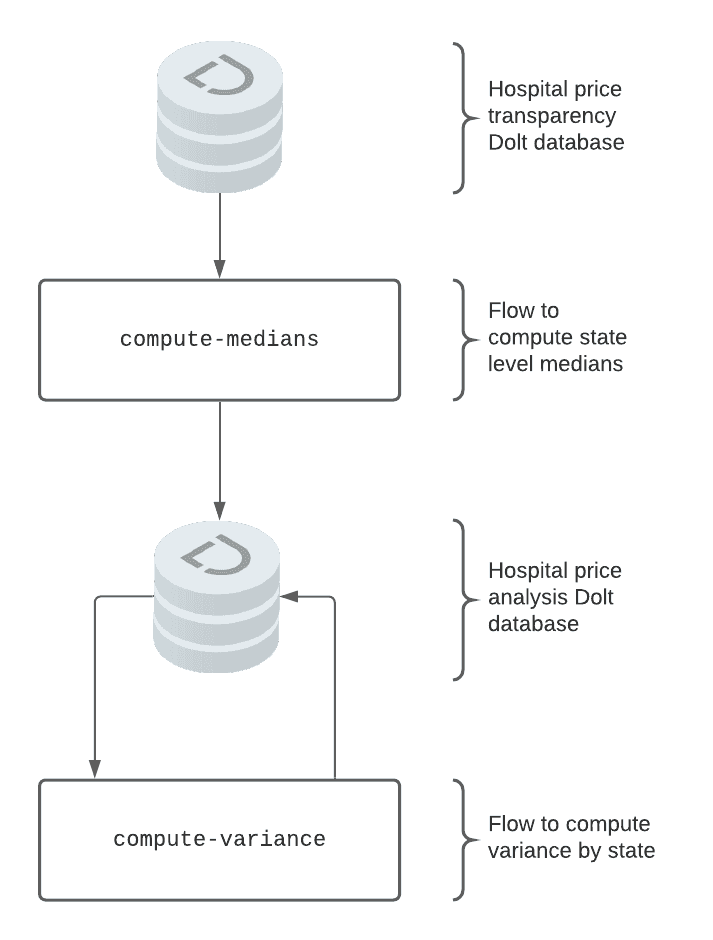
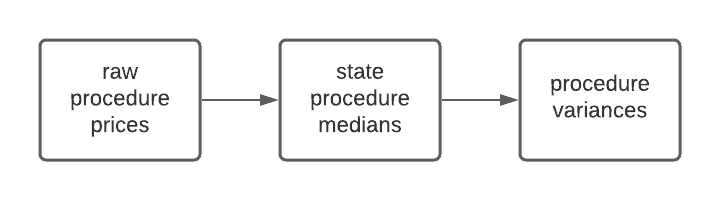
Our pipeline consists of two flows. One flow consumes the results written by the other. The first flow computes the state level median price for a hospital procedure. The second flow computes the variance of the price for a procedure across states. For input data we chose a new public dataset of 1400 US hospital prices created using DoltHub data bounties. Our pipeline will look something like this:
We will use the end result to illustrate how integrating Dolt and Metaflow provides Metaflow users with the ability to traverse versions of their final data, as well as traceback through the pipeline to examine various stages. Users can do this via the Metaflow API and Dolt integration in a familiar environment like a console or a notebook.
How it Works
Our design goal with this integration was to give the Metaflow user new capabilities directly from Metaflow while minimizing additional API surface area.
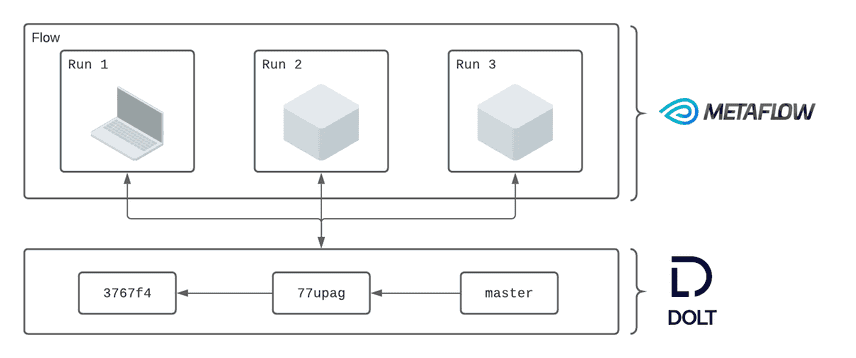
Workflows in Metaflow are called "flows." Each flow stores metadata about each execution, referred to as a "run." Each time a run interacts with Dolt it captures a small amount of metadata to make that interaction reproducible. We create a mapping between Metaflow runs and Dolt commits to provide users with powerful lineage and reproducibility features.
Runs of a flow read and write Pandas DataFrame objects to Dolt:
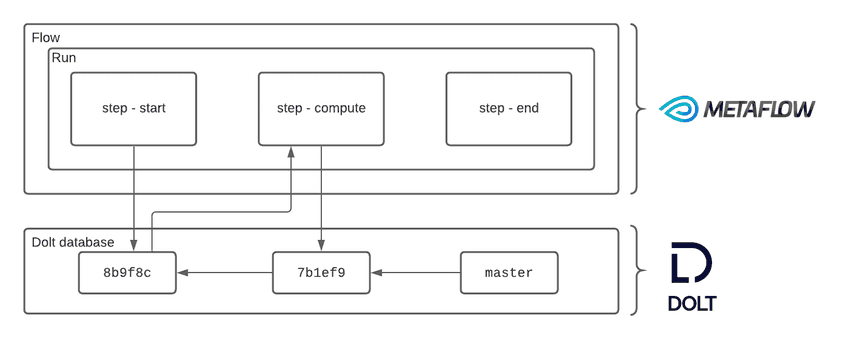
Individual steps of a run can create separate commits that snapshot the state of the database following a write:
When a flow reads data from Dolt, it records exactly how that data was read inside Metaflow. When a flow writes to Dolt it creates a commit and captures the metadata, as well as formatting the commit message. This allows users to browse the inputs and outputs of their flows from the Metaflow API directly without having to know much of anything about Dolt. Furthermore users can retrieve the flow that last touched a table at a given branch or commit, also directly from the Metaflow API.
This is all abstract, so let's install a few dependencies, grab a dataset, and get stuck into running our pipeline.
Setup
Let's get the boring stuff out of the way. We need the following:
- Dolt and
dolt-integrationsinstalled - Metaflow installed
- the sample dataset we will use, which can easily clone from DoltHub
Install Dolt
The first step is to install Dolt on a *nix system:
sudo bash -c 'curl -L https://github.com/dolthub/dolt/releases/latest/download/install.sh | bash'There are Windows distributions and a Homebrew cask. Find more details about installation here.
Install doltpy-integrations[metaflow]
Next let's install the Metaflow + Dolt integration. It comes packaged with both Metaflow, and Dolt's Python API, Doltcli. It's easy enough to install via pip:
pip install doltpy-integraions[metaflow]Get The Data
The final step is to acquire the dataset. Because Dolt is a SQL database with Git-like version control features, it includes the ability to clone a remote to your local machine. We can use that feature to easily acquire a Dolt database from a remote:
$ dolt clone dolthub/hospital-price-transparency && cd hospital-price-transparencyNote this dataset is nearly 20 gigabytes, and could take a few minutes to clone. Once it's landed it's straightforward to jump right into SQL:
At the outset we said we were going to store the results in Dolt. To do so we need to create a Dolt database to write to:
$ mkdir ~/hospital-price-analysis && cd ~/hospital-price-analysis
$ dolt init
Successfully initialized dolt data repository.We are now ready to start running our Metaflow based pipeline.
Using Metaflow
Before we get into the details, let's first produce a run of our pipeline off the latest version of the upstream database. The first flow computes the median cost of a given hospital procedure at the state level:
$ poetry run python3 hospital_procedure_price_state_medians.py run \
--hospital-price-db path/to/hospital-price-transparency \
--hospital-price-analysis-db path/to/hospital-price-analysis
Metaflow 2.2.8 executing HospitalPriceStateMedians for user:oscarbatori
Validating your flow...
The graph looks good!
2021-04-07 08:50:36.934 Workflow starting (run-id 1617810636925188):
2021-04-07 08:50:36.941 [1617810636925188/start/1 (pid 21075)] Task is starting.
2021-04-07 09:02:36.095 [1617810636925188/start/1 (pid 21075)] Task finished successfully.
2021-04-07 09:02:36.136 [1617810636925188/end/2 (pid 21257)] Task is starting.
2021-04-07 09:02:37.697 [1617810636925188/end/2 (pid 21257)] Task finished successfully.
2021-04-07 09:02:37.698 Done!The second flow computes the variance in median procedure price across states:
$ poetry run python3 hospital_procedure_price_variance_by_state.py run \
--hospital-price-analysis-db path/to/hospital-price-analysis
Metaflow 2.2.8 executing HospitalProcedurePriceVarianceByState for user:oscarbatori
Validating your flow...
The graph looks good!
2021-04-07 09:07:25.262 Workflow starting (run-id 1617811645255563):
2021-04-07 09:07:25.269 [1617811645255563/start/1 (pid 21299)] Task is starting.
2021-04-07 09:08:25.200 [1617811645255563/end/2 (pid 21349)] Task is starting.
2021-04-07 09:08:25.982 [1617811645255563/end/2 (pid 21349)] Task finished successfully.
2021-04-07 09:08:25.983 Done!We now have our first result set computed. Let's access the computed variances via the integration, using the flow as an entry point. You can execute the following snippet, and subsequent examples, using the Python interpreter on the command line or in a notebook. The snippet uses Metaflow’s Client API to access results of past runs:
from metaflow import Flow
from dolt_integrations.metaflow import DoltDT
dolt = DoltDT(run=Flow('HospitalProcedurePriceVarianceByState').latest_successful_run)
df = dolt.read('variance_by_procedure')
print(df)We see that we have successfully computed procedure level variance:
code price
0 CPT® 86708,72 253.125
1 CPT® 86708,73 253.125
2 CPT® 86708,74 253.125
3 CPT® 86708,75 253.125
4 CPT® 86708,76 253.125
... ...
1176694 nan,5 254148.840
1176695 nan,6 326868.470
1176696 nan,7 779888.100
1176697 nan,8 2152957.800
1176698 nan,9 4031522.000
[1176699 rows x 2 columns]We have seen it's relatively straightforward to run our pipeline, and access our versioned results via a reference to the flow that produced them. We now dive into some of the capabilities this provides Metaflow users who choose to use Dolt in their infrastructure.
Back-testing
In this example our input dataset is stored in Dolt. We used a DoltHub dataset because it's easy to clone the dataset and get started, and after all this post is about integrating Dolt with Metaflow. But having our input dataset in Dolt isn't just a matter of convenience for a demo. Because every Dolt commit represents the complete state of the database at a point in time, we can easily point our pipeline to historical versions of the data. Let's examine the Dolt commit graph and grab a commit straight from the SQL console:
$ cd path/to/hospital-price-transparency
$ dolt sql
# Welcome to the DoltSQL shell.
# Statements must be terminated with ';'.
# "exit" or "quit" (or Ctrl-D) to exit.
hospital_price_transparency> select commit_hash, message from dolt_commits where `date` < '2021-02-17' order by `date` desc limit 10;
+----------------------------------+------------------------------------------------------------+
| commit_hash | message |
+----------------------------------+------------------------------------------------------------+
| f0lecmblorr67rcuhuti6tbkriigh6gt | Updating prices with changes from uwmc_prices.csv |
| mj9ce6d8em9avj9ej0pqnaoes4fbglti | Updating cpt_hcpcs with changes from uwmc_cpt_hcpcs.csv |
| 2j6ommult20qvbj05j1nq63nkbd5fgdj | Updating prices with changes from prices.csv |
| pu8ctvhfcpp83q3iil8trp90vnuesaci | Updating cpt_hcpcs with changes from cpt_hcpcs.csv |
| q49l0kgnbbbgkt3imjd57tslbi2iges8 | Updating hospitals with changes from hospitals.csv |
| gstcq5loi9ieqdv1elrljab9hcgr090p | Updating hospitals with changes from hospitals.csv |
| te6spcqtjk0scose2c45f9t7tpcrt69c | Added hospital WellSpan Surgery & Rehabilitation Hospital. |
| bjg3b5lua8omadcl5nr6o7v0nphliqpu | Added hospital WellSpan York Hospital. |
| t4js1g5mfvgikqlmqa238it26mg94g5i | Added hospital Children's of Alabama. |
| jsan7p4iad61cjmeti858ebcl4s86vda | Added hospital McLaren Lapeer Region. |
+----------------------------------+------------------------------------------------------------+Suppose we'd like to run our pipeline with input data as of commit gstcq5loi9ieqdv1elrljab9hcgr090p, the first one labeled Updating hospitals with changes from hospitals.csv. That's easy enough, first let's name the commit with a branch:
$ dolt branch metaflow-backtest gstcq5loi9ieqdv1elrljab9hcgr090pBefore recomputing the medians let's create a branch in our hospital-price-analysis database associated with the root commit to store these experiments:
$ cd path/to/hospital-price-analysis
$ dolt branch
* master
metaflow-backtest
$ dolt checkout -b metaflow-backtest
Switched to branch 'metaflow-backtest'Now let's kick off recomputing the medians. Since we are recomputing our medians from a historical version of the raw pricing data we will write them to the separate experimentation branch we created above:
$ poetry run python3 hospital_procedure_price_state_medians.py run \
--hospital-price-db path/to/hospital-price-transparency \
--hospital-price-db-branch metaflow-backtest \
--hospital-price-analysis-db path/to/hospital-price-analysis \
--hospital-price-analysis-db-branch metaflow-backtest
Metaflow 2.2.8 executing HospitalPriceStateMedians for user:oscarbatori
Validating your flow...
The graph looks good!
2021-04-07 09:20:00.883 Workflow starting (run-id 1617812400875290):
2021-04-07 09:20:00.889 [1617812400875290/start/1 (pid 21700)] Task is starting.
2021-04-07 09:20:20.254 [1617812400875290/start/1 (pid 21700)] Task finished successfully.
2021-04-07 09:20:20.264 [1617812400875290/end/2 (pid 21776)] Task is starting.
2021-04-07 09:20:21.422 [1617812400875290/end/2 (pid 21776)] Task finished successfully.
2021-04-07 09:20:21.423 Done!And the variances can be run similarly, again using the experimentation branch:
$ poetry run python3 hospital_procedure_price_variance_by_state.py run \
--hospital-price-analysis-db path/to/hospital-price-analysis \
--hospital-price-analysis-db-branch metaflow-backtest
Metaflow 2.2.8 executing HospitalProcedurePriceVarianceByState for user:oscarbatori
Validating your flow...
The graph looks good!
2021-04-07 09:21:12.296 Workflow starting (run-id 1617812472287058):
2021-04-07 09:21:12.303 [1617812472287058/start/1 (pid 21827)] Task is starting.
2021-04-07 09:21:16.321 [1617812472287058/end/2 (pid 21871)] Task is starting.
2021-04-07 09:21:17.283 [1617812472287058/end/2 (pid 21871)] Task finished successfully.
2021-04-07 09:21:17.284 Done!We can now query the results directly from Python:
from metaflow import Flow
from dolt_integrations.metaflow import DoltDT
dolt = DoltDT(run=Flow('HospitalProcedurePriceVarianceByState').latest_successful_run)
df = dolt.read('variance_by_procedure')
print(df)Since this commit was taken from far earlier in the data gathering process, we can see far fewer unique procedure codes, 21K vs 1.17M:
code price
0 0001A 184.03088
1 0001M 12250.82000
2 0001U 35871.70000
3 0002A 275.67660
4 0002M 0.00000
... ...
21332 L1830-00 13.41620
21333 L1830-01 9.24500
21334 L1830-02 20.22480
21335 L1830-03 247.53125
21336 L1830-04 35.70125
[21337 rows x 2 columns]Or we can use Dolt SQL's AS OF syntax to query the results of our backtest:
$ cd path/to/hospital-price-analysis
$ dolt sql
# Welcome to the DoltSQL shell.
# Statements must be terminated with ';'.
# "exit" or "quit" (or Ctrl-D) to exit.
hospital_price_analysis> select * from variance_by_procedure as of 'metaflow-backtest' order by price desc limit 10;
+-------+---------------+
| code | price |
+-------+---------------+
| Q2042 | 5.9482256e+11 |
| Q2041 | 5.1718203e+11 |
| 216 | 1.2761694e+11 |
| J3399 | 9.192812e+10 |
| Q4142 | 1.604416e+10 |
| 0100T | 1.4931476e+10 |
| C9293 | 9.202785e+09 |
| 47133 | 8.558203e+09 |
| 90288 | 8.4152607e+09 |
| J7311 | 8.2143995e+09 |
+-------+---------------+In this section we saw how storing flow inputs in Dolt makes back-testing straightforward. Dolt's commit graph makes it easy to specify a historical database state.
Reproducibility
Our pipeline contains two steps, one computes state procedure price medians, and the second computes procedure price variances across states. Suppose now that we would like to tweak the way we compute variances. We might like to exclude some outliers, or invalid procedure codes. In a production setting we might own the variances computation but not the medians computation, and have stricter criteria for excluding invalid data. Let's update our variances job and then recompute using a fixed.
Let's first look at our Flow definition to exclude corrupt procedure codes:
@step
def start(self):
analysis_conf = DoltConfig(
database=self.hospital_price_analysis_db,
branch=self.hospital_price_analysis_db_branch
)
with DoltDT(run=self.historical_run_path or self, config=analysis_conf) as dolt:
median_price_by_state = dolt.read("state_procedure_medians")
clean = median_price_by_state[~median_price_by_state['code'].str.startswith('nan')]
variance_by_procedure = clean.groupby("code").var()
dolt.write(variance_by_procedure, "variance_by_procedure")
self.next(self.end)
Reproducibility comes from passing the path to a previous run:
historical_run_path = Parameter(
"historical-run-path", help="Read the same data as a path to a previous run"
)We use that path as a parameter to DoltDT, which in turn causes DoltDT to read data in exactly the same way as the run specified by the provided run path:
analysis_conf = DoltConfig(
database=self.hospital_price_analysis_db,
branch=self.hospital_price_analysis_db_branch
)
audit = Run(self.historical_run_path).data.dolt if self.historical_run_path else None
with DoltDT(run=self, config=analysis_conf, audit=audit) as dolt:Looking back to our first run of the flow computing the medians, the run ID was 1617810636925188. We can also see this in Dolt:
dolt log
commit lmjqpf293qg9ma10r034htt7jhfu5c4f
Author: oscarbatori <oscarbatori@gmail.com>
Date: Wed Apr 07 09:08:24 -0700 2021
Run: HospitalProcedurePriceVarianceByState/1617811645255563/start/1
commit 3je2okkrig5h3dbmuf8bhmfq0okps3lg
Author: oscarbatori <oscarbatori@gmail.com>
Date: Wed Apr 07 09:02:21 -0700 2021
Run: HospitalPriceStateMedians/1617810636925188/start/1Let's crate a branch to reproduce our run on pinned to the commit we want to reproduce from:
$ cd path/to/hospital-price-analysis
$ dolt branch
* master
metaflow-backtest
$ dolt branch metaflow-change 3je2okkrig5h3dbmuf8bhmfq0okps3lgWe can pass this branch straight into our job to achieve the desired data read isolation for testing our code changes:
$ poetry run python3 hospital_procedure_price_variance_by_state.py run \
--hospital-price-analysis-db ~/Documents/dolt-dbs/hospital-price-analysis \
--hospital-price-analysis-db-branch metaflow-change
Metaflow 2.2.8 executing HospitalProcedurePriceVarianceByState for user:oscarbatori
Validating your flow...
The graph looks good!
2021-04-07 15:11:15.634 Workflow starting (run-id 1617833475626845):
2021-04-07 15:11:15.641 [1617833475626845/start/1 (pid 42039)] Task is starting.
2021-04-07 15:12:17.799 [1617833475626845/start/1 (pid 42039)] Task finished successfully.
2021-04-07 15:12:17.808 [1617833475626845/end/2 (pid 42199)] Task is starting.
2021-04-07 15:12:19.046 [1617833475626845/end/2 (pid 42199)] Task finished successfully.
2021-04-07 15:12:19.047 Done!We can now see that our procedure level variances have been filtered appropriately and we no longer have corrupt procedure codes in our dataset, first grabbing it using the run path:
from dolt_integrations.metaflow import DoltDT
from metaflow import Run
doltdt = DoltDT(run=Run('HospitalProcedurePriceVarianceByState/1617833475626845'))
df =doltdt.read('variance_by_procedure')And we have eliminated the corrupt procedure codes:
index code price
0 679855 CPT® 78710,103 0.000
1 679856 CPT® 78710,104 0.000
2 679857 CPT® 78710,105 0.000
3 679858 CPT® 78710,106 0.000
4 679859 CPT® 78710,107 0.000
... ... ...
1176681 2190407 HCPCS C1769,15784 5886.125
1176682 2190408 HCPCS C1769,15785 5886.125
1176683 2190409 HCPCS C1769,15786 5886.125
1176684 2190410 HCPCS C1769,15787 5886.125
1176685 2190411 HCPCS C1769,15788 5886.125
[1176686 rows x 3 columns]By simply retrieving a run path, and kicking off our variances flow, we were able to reproduce the exact inputs of a historical run.
Lineage
In the previous section we showed how to run one Flow using the inputs of a previous run. We did this to achieve data version isolation for the purposes of testing our code changes. The same mechanism we used for achieving this kind of reproducibility also allows us to track data lineage. Recall that our pipeline has two steps, each a separate flow:
Obviously a real world example might have a much more complicated data dependency graph, making this kind of tracking all the more important. Let's see how we would trace the lineage of the final variances. The first thing to do is grab the run that created the current production data:
from dolt_integrations.metaflow import DoltDT, DoltConfig
downstream_doltdt = DoltDT(config=DoltConfig(database='/Users/oscarbatori/Documents/dolt-dbs/hospital-price-analysis'))
run = downstream_doltdt.get_run('variance_by_procedure')
print(run)Which produces a Metaflow run path:
'HospitalProcedurePriceVarianceByState/1617811645255563/start/1'We now have the run ID of the flow that produced the medians from which we computed our variances. We can access the flow that wrote the medians in a similar manner:
commit = Run('HospitalProcedurePriceVarianceByState/1617811645255563').data.dolt['actions']['state_procedure_medians']['commit']
run = downstream_doltdt.get_run('state_procedure_medians', commit)
print(run)And we have traced back to the medians job that originally produced this data:
'HospitalPriceStateMedians/1617810636925188/start/1'As a final step we can grab the input data, since we stored in Dolt:
upstream_doltdt = DoltDT(run=Run('HospitalPriceStateMedians/1617810636925188'))
df = upstream_doltdt.read('prices')
print(df)Which yields the original input dataset we started out with:
Out[10]:
npi_number ... price
0 1053358010.0 ... 75047.00
1 1336186394 ... 75047.00
2 1003139775.0 ... 457.23
3 1053824292.0 ... 972.00
4 1417901406.0 ... 296.00
... ... ...
72724847 1184897647 ... 3724.00
72724848 1003858408 ... 96.11
72724849 1003858408 ... 94.53
72724850 1598917866 ... 22.00
72724851 1598917866 ... 15.40
[72724852 rows x 4 columns]By storing Metaflow results in Dolt, result sets can be associated with flows and the input datasets. When results from Metaflow are put into other data stores we don't have a way to trace the table back to flow run that produced it.
Conclusion
In this post we demonstrated how to use Dolt alongside Metaflow. Metaflow provides a framework for defining data engineering and data science workflows. Using Dolt for inputs and outputs augments pipelines defined in Metaflow with additional capabilities. Users can examine a table in their Dolt database and locate the flow that produced that table, and if that flow used Dolt as an input, locate the flows that rate the input data, and so on. Users can also run a flow pinning a historical version of the data, providing for reproducible runs that use data version isolation to ensure code changes are properly tested. Finally, when Dolt is used as an input the commit graph can be used for back-testing against historical versions of the data.